HiCExplorer article comes online in nature communications
Our article on DNA sequence behind Drosophila (Fly) genome architecture is now published in Nature Communications. The preprint has already been available on biorXiv since march 2017.
In this article, we introduce HiCExplorer: An easy to use tool for HiC data analysis, also available in galaxy. We also introduce HiCBrowser : A standalone software to visualize HiC along with other genomic datasets. Based on HiCExplorer and HiCBrowser, we built a useful resource for anyone to browse and download the chromosome conformation datasets in Human, Mouse and Flies. It’s called the chorogenome navigator.
Along with these resources, we present an analysis of DNA sequences behind 3D genome of Flies. Using high-resolution HiC analysis, we find a set of DNA motifs that characterize TAD boundaries in Flies and show the importance of these motifs in genome organization.
Below is figure 1 from our paper, denoting a region in drosophila genome with TADs, associated TAD-score (from hicexplorer), boundary associated proteins and histone marks. Most boundaries are in active regions of the genome and associate with promoters of constituively expressed genes. Active TADs are the smallest in size on average.
We hope that our analysis and these resources presented here would be useful for the community and welcome any feedback.
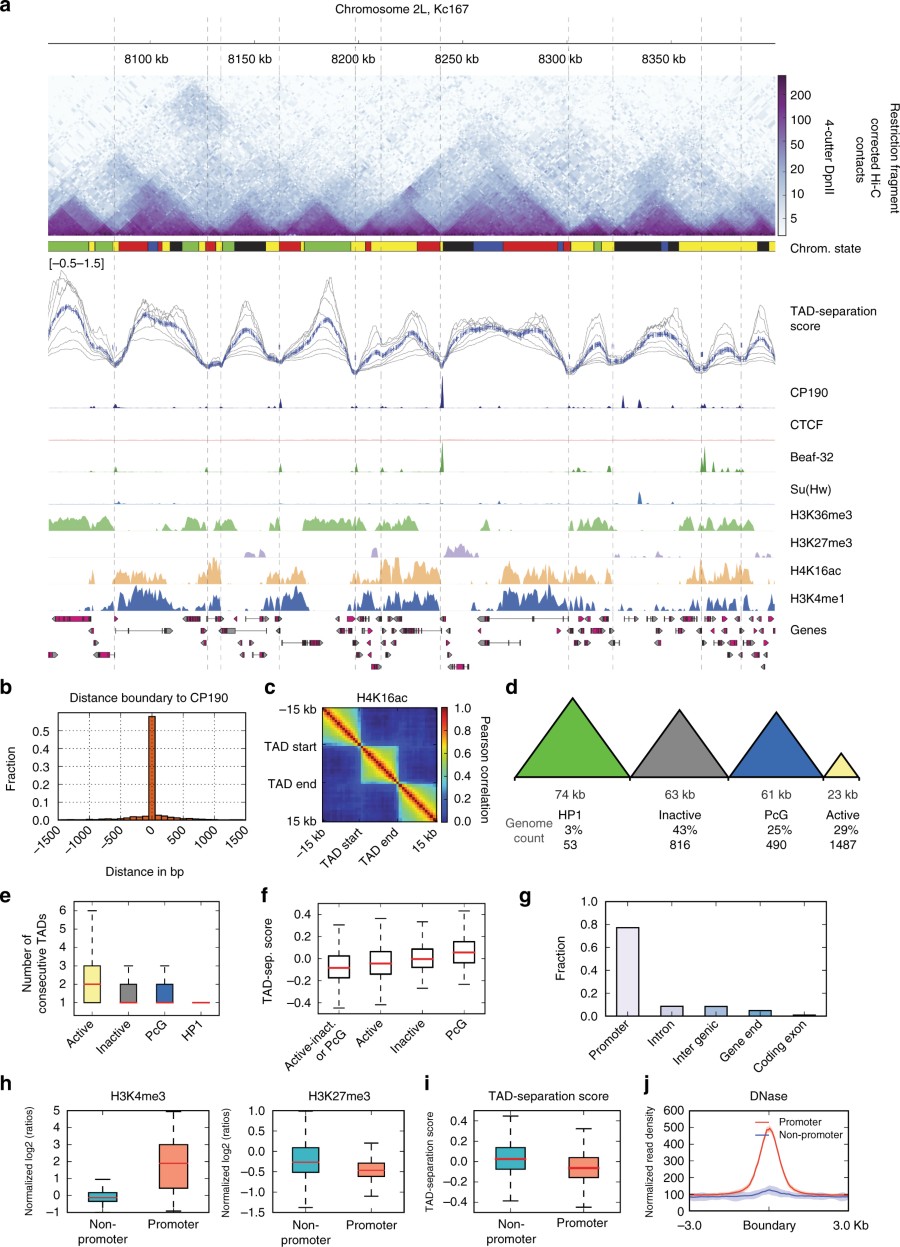
figure 1